Retrosynthetically Decompose a Molecule In a Tree Structure Using Recap in RDKit
Retrosynthetic analysis involves decomposing a target molecule into a set of fragments that could be combined to make the parent molecule using common reactions. The Recap algorithm by X. Lewell, D. Judd, S. Watson, and M. Hann accomplishes that. Recap is implemented in the RDKit cheminformatics Python package.
Download this notebook from GitHub by right-clicking and choosing Save Link As…
RDKit helpfully provides a RecapHierarchyNode structure of nodes, where the keys are SMILES strings corresponding to the fragment, and the values are nodes containing child fragments. However, it is not easy to visualize the results, because the results are SMILES strings and the hierarchy is not shown visually.
This utility function molecule_recap_tree allows you to visualize both the fragments and their hierarchy. Here is an example, of [6-(Hydroxymethoxy)pyridin-2-yl]oxymethanol, annotated to explain the hierarchy:
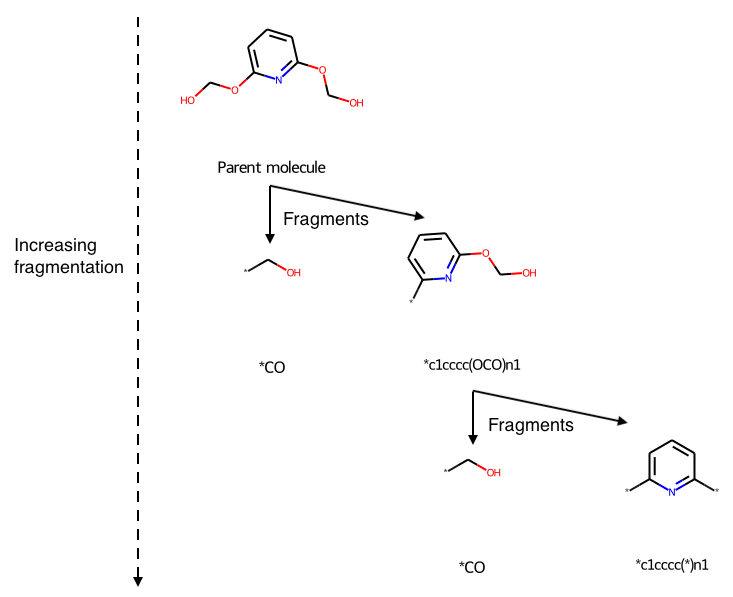
As compared to the same molecule using the RecapDecompose function directly, showing only the immediate children:
from rdkit import Chem
from rdkit.Chem import Draw, Recap
molecule = Chem.MolFromSmiles("n1c(OCO)cccc1-OCO")
hierarch = Recap.RecapDecompose(molecule)
ks=hierarch.children.keys()
sorted(ks)
['*CO', '*c1cccc(OCO)n1']
def molecule_recap_tree(smiles:str, name:str="", verbose=False, **kwargs):
"""
Draw a molecular fragmentation tree using the Recap algorithm given in
https://www.semanticscholar.org/paper/RECAP-%E2%80%94-Retrosynthetic-Combinatorial-Analysis-A-New-Lewell-Judd/fbfb10d1f63aa803f6d47df6587aa0e41109f5ee
:returns: RDKit grid image, and (if verbose=True) RDKit molecule of parent molecule, Recap hierarchy, nonbinary tree hierarchy, and fragment grid
:rtype: RDKit grid image, and (if verbose=True) rdkit.Chem.rdchem.Mol, rdkit.Chem.Recap.RecapHierarchyNode, NonBinTree, list[list[str]]
:param smiles: The SMILES string of the molecule to be fragmented
:param name: The name of the parent molecule; if not supplied, it will be labled with its SMILES string
:param verbose: Whether to return verbose output; default is False so calling this function will present a grid image automatically
"""
molecule = Chem.MolFromSmiles(smiles)
RecapHierarchy = Recap.RecapDecompose(molecule)
root = NonBinTree(RecapHierarchy.smiles)
molecule_nonbinary_tree = get_children(RecapHierarchy, root)
fragment_grid = molecule_nonbinary_tree.get_grid()
mols_per_row = len(fragment_grid[0])
recap_plot = [item for sublist in fragment_grid for item in sublist]
recap_labels = [item for sublist in fragment_grid for item in sublist]
if name:
recap_labels[0] = name
# Use MolsMatrixToGridImage if available in the installed version of RDKit
molsMatrix = [[Chem.MolFromSmiles(smile) for smile in sublist] for sublist in fragment_grid]
try:
drawing = Draw.MolsMatrixToGridImage(molsMatrix=molsMatrix, legendsMatrix=fragment_grid, **kwargs)
except AttributeError:
drawing = Draw.MolsToGridImage([Chem.MolFromSmiles(smile) for smile in recap_plot], legends=recap_labels, molsPerRow=mols_per_row, **kwargs)
if verbose:
return drawing, molecule, RecapHierarchy, molecule_nonbinary_tree, fragment_grid
else:
return drawing
def concat_grids_horizontally(grid1:list[list[str]], grid2:list[list[str]]) -> list[list[str]]:
"""Concatenate two nested lists horizontally, for example
inputs [['a'],['b'],['c']] and [['d'], ['e'], ['f']]
produce [['a', 'd'], ['b', 'e'], ['c', 'f']]
:returns: The combined grid, a two-deep nested list of strings
:param grid1: The first grid, a two-deep nested list of strings
:param grid2: The second grid, a two-deep nested list of strings
"""
if grid1 == [[]]:
combined = grid2
elif grid2 == [[]]:
combined = grid1
else:
combined = []
for row_counter in range(len(grid1)):
combined += [grid1[row_counter] + grid2[row_counter]]
return combined
class NonBinTree:
"""
Nonbinary tree class
Note that this class is not designed to sort nodes as they are added to the tree;
the assumption is that they should be ordered in the order added
Adapted from https://stackoverflow.com/questions/60579330/non-binary-tree-data-structure-in-python#60579464
"""
def __init__(self, val:str):
"""Create a NonBinTree instance"""
self.val = val
self.nodes = []
def add_node(self, val:str):
"""Add a node to the tree and return the new node"""
self.nodes.append(NonBinTree(val))
return self.nodes[-1]
def __repr__(self) -> str:
"""Print out the tree as a nested list"""
return f"NonBinTree({self.val}): {self.nodes}"
def get_ncols(self) -> int:
"""Get the number of columns in the tree"""
self.ncols = 0
if len(self.nodes) > 0:
# If there are nodes under this one, call get_ncols on them recursively
for node in self.nodes:
self.ncols += node.get_ncols()
else:
# If there are no nodes under this one, add 1 for this node
self.ncols += 1
return self.ncols
def get_max_depth(self) -> int:
"""Get the maximum depth of the tree"""
max_depth = 0
if len(self.nodes) > 0:
for node in self.nodes:
this_depth = node.get_max_depth()
max_depth = max(this_depth + 1, max_depth)
else:
max_depth = max(1, max_depth)
self.max_depth = max_depth
return self.max_depth
def get_grid(self) -> list[list[str]]:
"""
Get a two-dimensional grid where
each row is a level in the fragment hierarchy, and
the columns serve to arrange the fragments horizontally
"""
# Call methods to calculate self.ncols and self.max_depth
self.get_ncols()
self.get_max_depth()
# Create top row: Node value, then the rest of columns are blank (empty strings)
grid = [[self.val] + [""] * (self.ncols - 1)]
n_nodes = len(self.nodes)
if n_nodes > 0:
nodes_grid = [[]]
# Iterate through the child nodes
for node_counter, node in enumerate(self.nodes):
# Recursively call this function to get the grid for children
node_grid = node.get_grid()
# Add spacer rows if needed
node_grid_rows = len(node_grid)
rows_padding = self.max_depth - node_grid_rows - 1
for padding in range(rows_padding):
node_grid += [[""] * len(node_grid[0])]
nodes_grid = concat_grids_horizontally(nodes_grid, node_grid)
grid += nodes_grid
return grid
def get_children(base_node:Chem.Recap.RecapHierarchyNode, root:NonBinTree = None) -> NonBinTree:
"""
Convert an RDKit RecapHierarchyNode into a NonBinTree by
traversing the RecapHierarchyNode, getting all its children recursively, and adding them to a NonBinTree
:returns: NoBinTree containing the Recap hierarchy
:param base_node: The RDKit RecapHierarchyNode
:param root: The NoBinTree containing only the root node
"""
for smiles, node in base_node.children.items():
added_tree_node = root.add_node(smiles)
children = node.children.keys()
# Sort the children nodes to get consistent ordering
children = sorted(children)
if len(children) > 0:
get_children(node, added_tree_node)
return root
Here is the unannotated output:
molecule_recap_tree("n1c(OCO)cccc1-OCO", "Parent molecule")
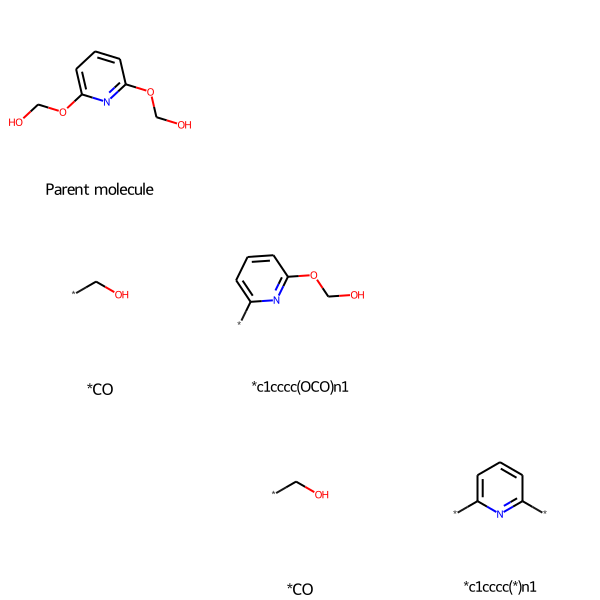
Stars (“dummy atoms”) show points where the molecule was fragmented.
The key RDKit commands that molecule_recap_tree uses are:
RecapDecomposeto decompose the parent molecule into successive fragmentsMolsToGridImageto draw the fragment hierarchy in a grid, and label the fragments with their SMILES strings
Get Additional Data
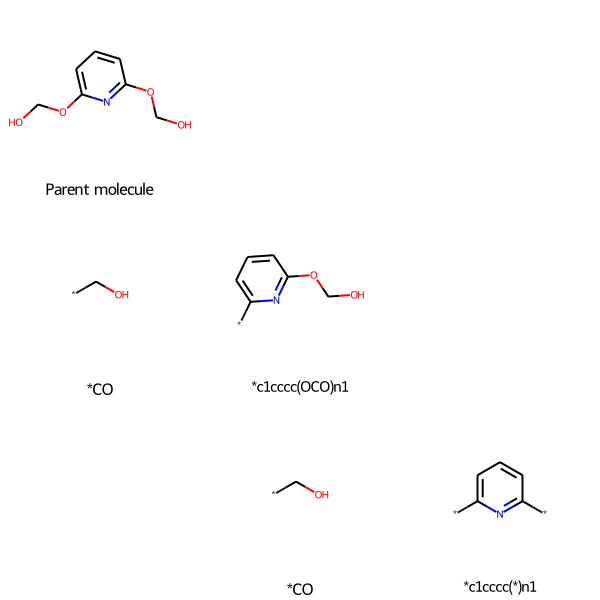
If you want molecule_recap_tree to return not just the grid image, but also the parent molecule, the RDKit Recap hiearchy node, the non-binary tree hierarchy, and grid of fragment hierarchy, set verbose=True:
drawing, molecule, RecapHierarchy, molecule_nonbinary_tree, fragment_grid = molecule_recap_tree("n1c(OCO)cccc1-OCO", "Parent molecule", verbose=True)
You then must explicitly call the hierarchy image to draw it:
drawing
molecule is the RDKit parent molecule:
molecule
RecapHierarchy is the RDKit Recap hierarchy, which returns the top-level node, which you could traverse to obtain the hierarchy:
RecapHierarchy
<rdkit.Chem.Recap.RecapHierarchyNode at 0x112663cd0>
molecule_nonbinary_tree is the NonBinTree hierarchy, which contains the same hierarchy as the RecapHierarchy, and gives all the nodes directly:
molecule_nonbinary_tree
NonBinTree(OCOc1cccc(OCO)n1): [NonBinTree(*CO): [], NonBinTree(*c1cccc(OCO)n1): [NonBinTree(*CO): [], NonBinTree(*c1cccc(*)n1): []]]
fragment_grid is the two-dimensional fragment grid, where each row is a level in the fragment hierarchy, and the columns serve to arrange the fragments horizontally:
fragment_grid
[['OCOc1cccc(OCO)n1', '', ''],
['*CO', '*c1cccc(OCO)n1', ''],
['', '*CO', '*c1cccc(*)n1']]
Or, more nicely formatted into columns and with empty strings not shown:
for smiles1, smiles2, smiles3 in fragment_grid:
print (f"{smiles1:^20}{smiles2:^20}{smiles3:^20}")
OCOc1cccc(OCO)n1
*CO *c1cccc(OCO)n1
*CO *c1cccc(*)n1
Pass Arguments to MolsToGridImage
More complex molecules can easily produce many fragments:
molecule_recap_tree("Clc1cc(c(OC)cc1N)C(=O)NC3CCN(CCCOc2ccc(F)cc2)CC3OC", "parent molecule")
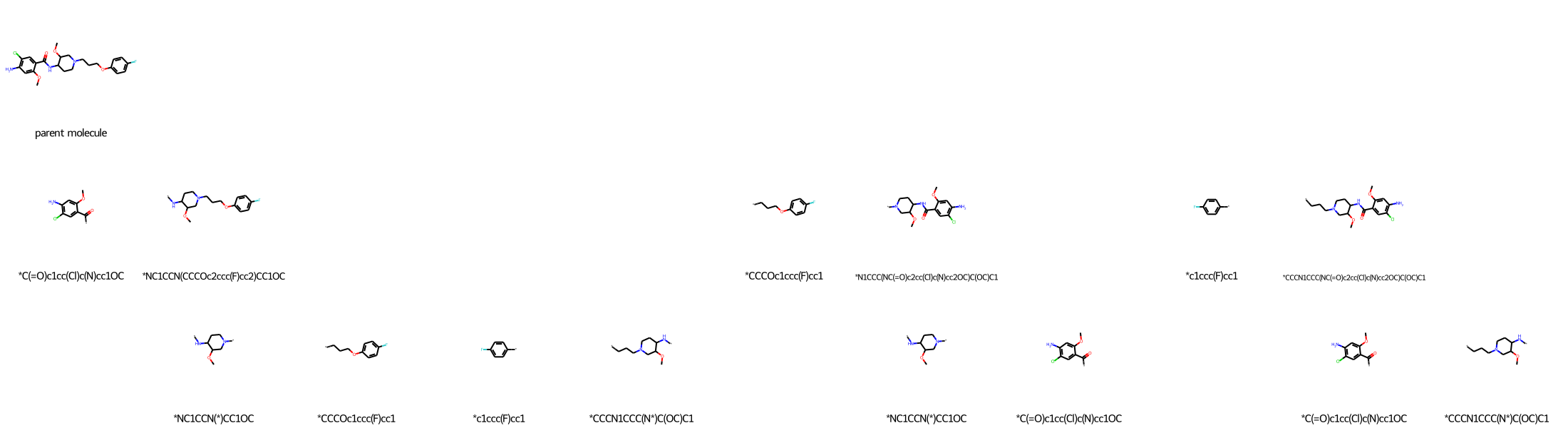
If you zoom into a fragment, you will notice that it is fuzzy due to the low resolution.
To address this case, you can use molecule_recap_tree’s ability to pass any keyword arguments of Draw.MolsToGridImage to that function. Specifically, you can pass the useSVG=True flag so that the molecular images do not lose resolution when you zoom into them. If you zoom into this image, you will notice that the fragments are easy to read because Scalable Vector Graphics is, as the name states, a vector format. (useSVG=True is not the default in molecule_recap_tree because bonds are not nicely joined as they are in the default PNG format.)
molecule_recap_tree("Clc1cc(c(OC)cc1N)C(=O)NC3CCN(CCCOc2ccc(F)cc2)CC3OC", "parent molecule", useSVG=True)
Note About Hierarchies
Another approach I could have taken was to extend the class rdkit.Chem.Recap.RecapHierarchyNode, rather than use the NonBinaryTree class. Using the NonBinaryTree class allows for different kinds of hierarchies to be depicted as a grid, as I demonstrate in other posts Visualizing Nonbinary Trees: Classification of Chemical Isomers and Draw a Mass Spectrometry Fragmentation Tree Using RDKit.
