Drawing molecules with EPAM’s Indigo cheminformatics library
Exploring other cheminformatics toolkits besides the RDKit, I wanted to try EPAM Indigo Toolkit. The Indigo Toolkit is free and open-source with Apache License 2.0, so it can be used in proprietary software. I was unable to find simple examples of drawing molecules in a Python Jupyter Notebook, so here’s how to do that. This post also demonstrates how to save molecular images to a file.
In general the official documentation for Indigo is much less than for the RDKit, as are the examples posted by users online. As a form of documentation, below I specify Indigo argument names, and am more pedantic about including code blocks and outputs.
Open this notebook in Google Colab so you can run it without installing anything on your computer
Import and initialize Indigo modules
Indigo works a little differently than the RDKit: With Indigo, you start by creating an instance of the class for Indigo or its renderer; then you can use the class’s methods.
from indigo import Indigo
from indigo.renderer import IndigoRenderer
# Initialize Indigo and IndigoRenderer
indigo = Indigo()
renderer = IndigoRenderer(indigo)
To display the images in a Jupyter Notebook, we use IPython display modules.
from IPython.display import display, SVG, Image
Set an Indigo option for all images
As a first example of performing an operation in Indigo, let’s set a drawing option that will apply to the rest of this post. This option, render-margins, sets the “Horizontal and vertical margins around the image, in pixels.”
indigo.setOption("render-margins", 10, 10)
It’s helpful to have such fine control of drawing. Compared to the RDKit, I found Indigo’s syntactical structure less transparent because you supply the option name in quotes, meaning you don’t get a list of parameter name options, or autocomplete in an IDE (integrated development environment).
Draw a single molecule with Indigo Renderer
Set up a single molecule
One nice feature of Indigo is that it will convert from a chemical name to a molecule, so you don’t need to compose the SMILES.
name = "3-ethyl-octane"
mol = indigo.nameToStructure(name)
Once you have the molecule, you can get the SMILES using the molecule’s smiles method.
mol.smiles()
'CCC(CCCCC)CC'
Display an Indigo molecular structure in the notebook
I was able to render a molecular structure in a Jupyter Notebook using either PNG or SVG format. In each code block, we start by setting the render-output-format option to the desired graphics format. The Jupyter Notebook command to display the image varies depending on the image format.
indigo.setOption("render-output-format", "png")
img = renderer.renderToBuffer(
obj=mol,
)
display(Image(img))
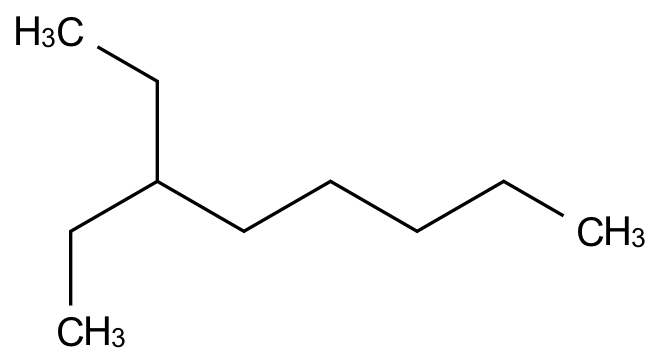
indigo.setOption("render-output-format", "svg")
mol_svg = renderer.renderToBuffer(
obj=mol,
)
display(SVG(mol_svg))
Taking a different approach than the RDKit, Indigo renders terminal carbon atoms explicitly (CH3 here) rather than implicitly as vertices.
Write an Indigo molecular structure to a file
Similarly, we can save a molecular structure to a file in either PNG or SVG format.
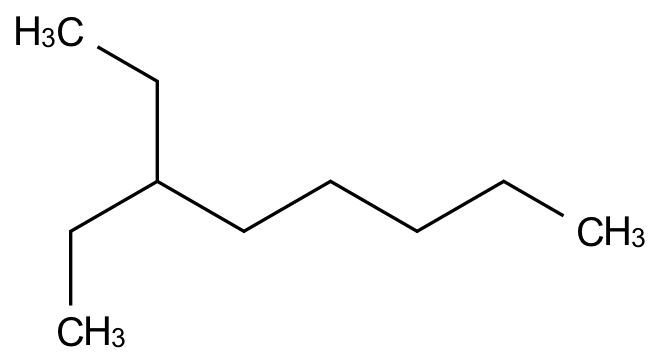
indigo.setOption("render-output-format", "png")
mol.layout()
renderer.renderToFile(
obj=mol,
filename="/images/indigo/mol.png",
)
Here’s the image in the PNG file:
indigo.setOption("render-output-format", "svg")
mol.layout()
renderer.renderToFile(
obj=mol,
filename="/images/indigo/mol.svg",
)
And the SVG file has the same molecular structure:
Draw a grid of molecules with Indigo Renderer
Often we want to draw a grid of molecular structures to show similarities amongst them, depict the results of a query, etc.
Set up an array of Indigo molecules
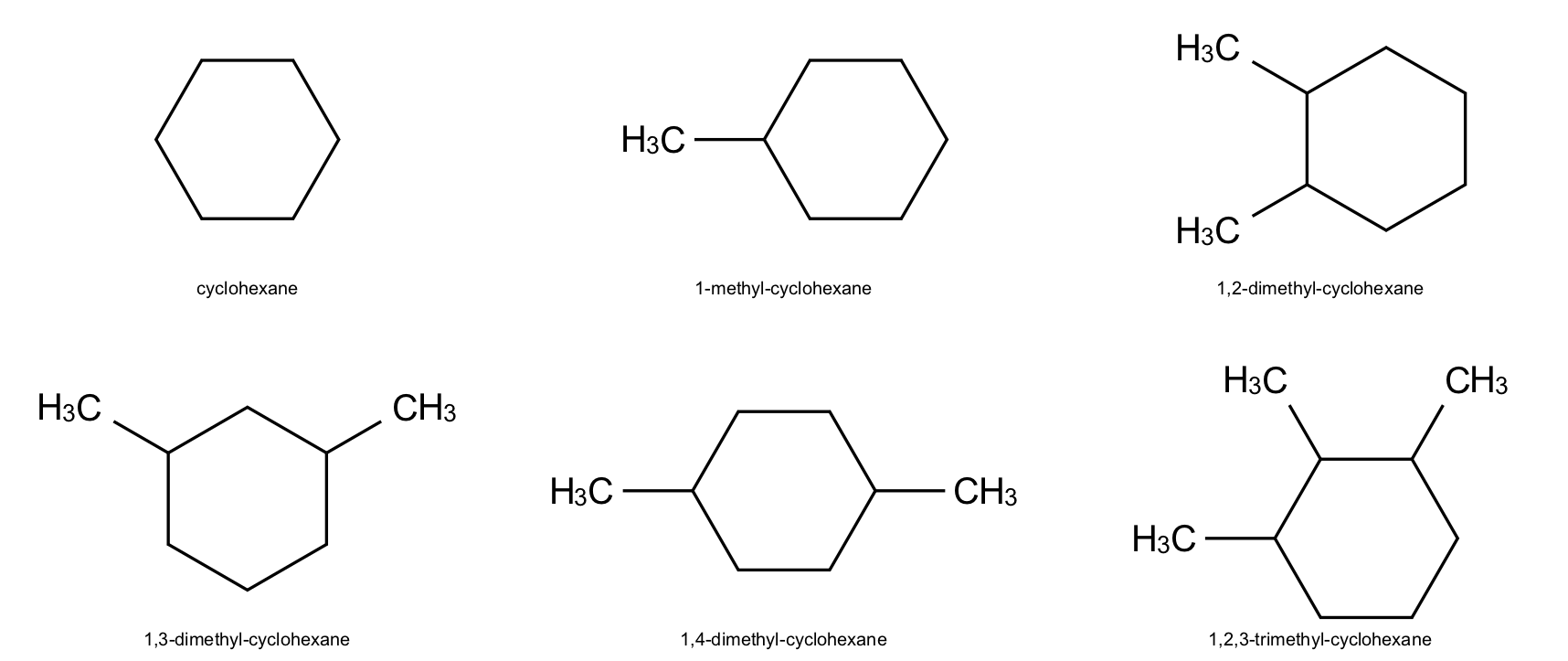
First let’s create a list of molecules based on cyclohexane.
names = [
"cyclohexane",
"1-methyl-cyclohexane",
"1,2-dimethyl-cyclohexane",
"1,3-dimethyl-cyclohexane",
"1,4-dimethyl-cyclohexane",
"1,2,3-trimethyl-cyclohexane",
]
In Indigo, we use createArray() to create an array of molecules to later draw in a grid. Let’s set the property grid-comment of each molecule to its name so we can later display its name in the drawing grid.
array = indigo.createArray()
for n in names:
this_mol = indigo.nameToStructure(n)
this_mol.layout()
this_mol.setProperty("grid-comment", n)
array.arrayAdd(this_mol)
In the options below, title refers to the label under each molecule in the grid. The title will be the molecule’s name because that’s what we previously set grid-comment to. Again, the granular ability to set specific layout properties, such as vertical and horizontal spacing, is nice.
# Set the label under each molecule
indigo.setOption("render-grid-title-property", "grid-comment")
# Set the vertical offset of the label under each molecule
indigo.setOption("render-grid-title-offset", "15")
# Set the spacing between grid cells as horizontal and vertical pixels
indigo.setOption("render-grid-margins", "70, 70")
Display a grid of Indigo molecular structures in the notebook as a PNG
Now we can draw a grid of molecules using renderGridToBuffer. We pass the array of molecules as the objects parameter. Similar to the RDKit, we can specify the number of columns, here with ncolumns. The parameter refatoms can be used to align structures to have the same orientation, though the code below doesn’t set it to anything.
indigo.setOption("render-output-format", "png")
img_grid_png = renderer.renderGridToBuffer(
objects=array,
refatoms=None,
ncolumns=3,
)
display(Image(img_grid_png))
Display a grid of Indigo molecular structures in the notebook as an SVG
The output from this has been intermittently problematic: the molecular structures have come out distorted, perhaps due to some issue with writing or displaying the SVG format. The SVG format specifies graphical elements and their locations, which has the advantages of vector graphics but this possible issue. On the other hand, PNG is a raster-graphics format where the image is specified as a grid of pixels, so it’s not surprising that it doesn’t have the same issue.
indigo.setOption("render-output-format", "svg")
img_grid = renderer.renderGridToBuffer(
objects=array,
refatoms=None,
ncolumns=3,
)
display(SVG(img_grid))
Write a grid of Indigo molecules to an image file
Both PNG and SVG formats are possible.
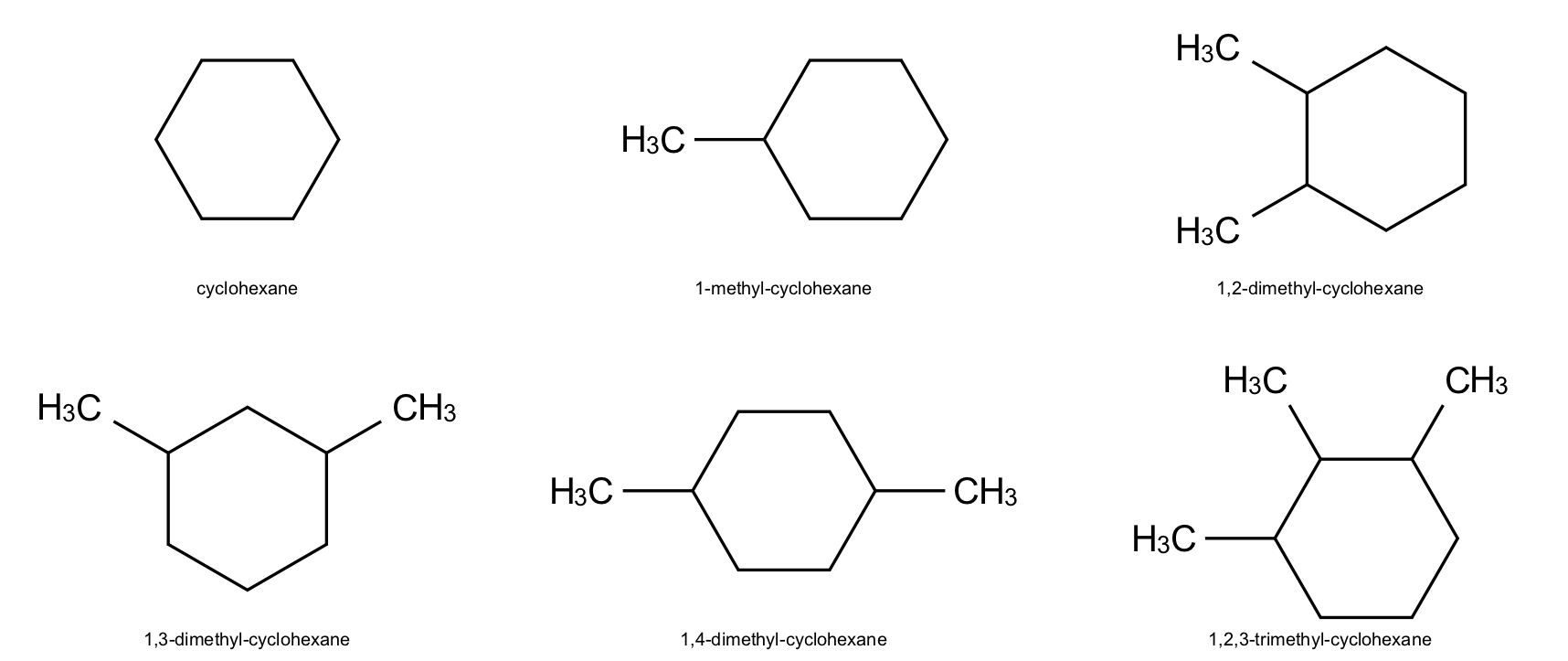
indigo.setOption("render-output-format", "png")
renderer.renderGridToFile(
objects=array,
refatoms=None,
ncolumns=3,
filename="/images/indigo/structures.png",
)
Here’s the image in the PNG file:
indigo.setOption("render-output-format", "svg")
renderer.renderGridToFile(
objects=array,
refatoms=None,
ncolumns=3,
filename="/images/indigo/structures.svg",
)
Here’s the image in the SVG file:
Another resource
You can find other examples of how to use Indigo at Chemistry Toolkit Rosetta Wiki, which has code examples for common operations in cheminformatics toolkits such as Indigo, RDKit, CDK, OpenBabel, OpenEye, CACTVS, Cinfony, and Chemkit. The page Depict a compound as an image is relevant to the goal of this post.



