Tutorial for Skeletal Editing in Drug Discovery
This notebook is the result of a collaboration between myself, Dr. Jeremy E. Monat, and Dr. Phyo Phyo Zin. This is a hands-on tutorial that guides readers through implementing skeletal editing transformations using Python and RDKit, and Phyo Phyo focuses on the high-level principles and applications in the blog post How to Use Skeletal Editing in Drug Discovery: Key Benefits & Applications. Together, our goal is to provide both a conceptual understanding and practical approach to this emerging field.
Open this notebook in Google Colab so you can run it without installing anything on your computer
Code foundation
First we install the RDKit.
%%capture
!pip install rdkit
Next we import the necessary modules.
from rdkit import Chem
from rdkit.Chem import AllChem
from rdkit.Chem.Draw import IPythonConsole
from IPython.display import display
Now we set up drawing options for the blog post.
IPythonConsole.molSize = 400, 200
Let’s define a function to display and run a reaction with RunReactants using the RDKit’s Reaction SMARTS format.
def run_rxn(rxn_sml, mol):
rxn = AllChem.ReactionFromSmarts(rxn_sml)
print("Reaction:")
display(rxn)
try:
products = rxn.RunReactants((mol,))
except IndexError:
raise IndexError(
"Reaction failed; the starting material may not match the reaction SMARTS"
)
return products
Now let’s fold that into a larger function that also displays the reactant and distinct products. Because of the way SMARTS matches parts of a molecule, we might have multiple identical products, so we only want to show each product once.
def plot_rxn(rxn_sml, mol):
try:
products = run_rxn(rxn_sml, mol)
except IndexError as e:
print(e)
return
print("Reactant:")
display(mol)
if not products:
print("No products")
return
print("Distinct products:")
product_smls = set()
for product in products:
product_mol = product[0]
Chem.SanitizeMol(product_mol)
# Get the canonical SMILES string of the product molecule
this_sml = Chem.MolToSmiles(product_mol)
# If this product has already been made, don't display it again
if this_sml in product_smls:
continue
product_smls.add(this_sml)
print(Chem.MolToSmiles(product_mol))
display(product_mol)
return product_smls
Skeletal Editing Reactions Used in Drug Discovery Blog Post
Here are three skeletal editing reactions used in Phyo Phyo’s blog post. In each case, we give
- the general reaction and apply it to a generic reactant, then
- a modified reaction and apply it to a molecule related to drug discovery.
Converting quinolines to quinazolines
This reaction is from Woo et al., 2023, scheme 1d.
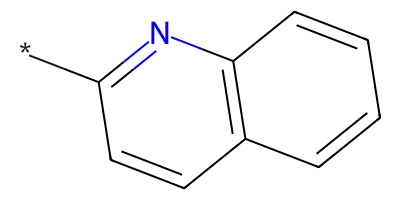
quinoline = "*c1nc2c(cccc2)cc1"
quinoline_mol = Chem.MolFromSmiles(quinoline)
quinoline_mol
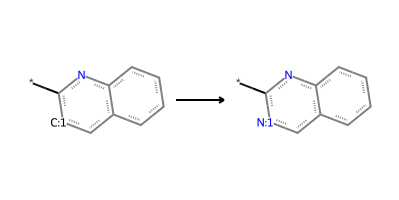
quinoline_to_quinazoline = "*-c1nc2c(cccc2)c[c:1]1>>*-c1nc2c(cccc2)c[n:1]1"
product_smls = plot_rxn(quinoline_to_quinazoline, quinoline_mol)
Reaction:
Reactant:
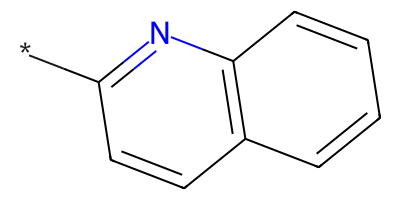
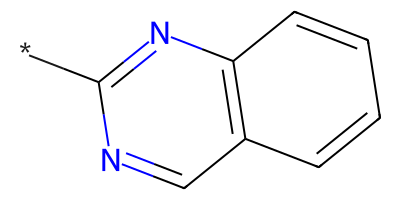
Distinct products:
*c1ncc2ccccc2n1
Skeletal Editing for Talnetant
quinoline_to_quinazoline_modified = "[cX3:1]1[n:2][c:3]2[c:4]([c:5][c:6][c:7][c:8]2)[c:9][c:10]1>>[cX3:1]1[n:2][c:3]2[c:4]([c:5][c:6][c:7][c:8]2)[c:9][n:10]1"
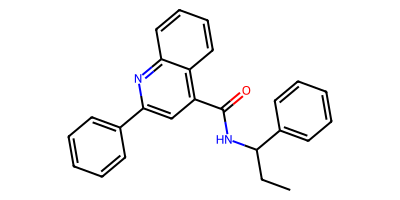
talnetant_smi = "c1ccccc1c2cc(C(=O)NC(CC)c4ccccc4)c3ccccc3n2"
talnetant_mol = Chem.MolFromSmiles(talnetant_smi)
product_smls = plot_rxn(quinoline_to_quinazoline_modified, talnetant_mol)
Reaction:
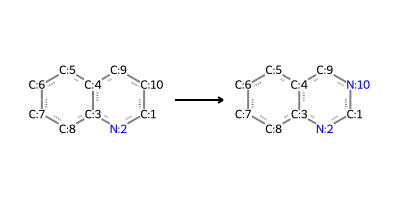
Reactant:
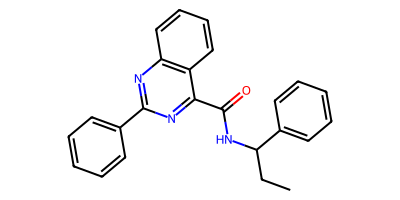
Distinct products:
CCC(NC(=O)c1nc(-c2ccccc2)nc2ccccc12)c1ccccc1
Converting pyrrolidines to cyclobutanes
This reaction is from Hui et al., 2021, Scheme 1.
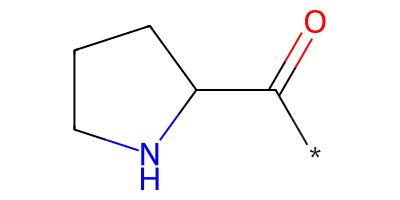
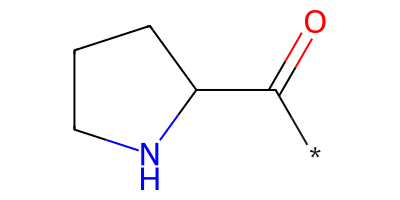
pyrrolidine = "*C(=O)C1CCCN1"
pyrrolidine_mol = Chem.MolFromSmiles(pyrrolidine)
pyrrolidine_mol
pyrrolidine_to_cyclobutane = "*-C(=O)-[C:1]1-C-C-[C:2]-N-1>>*-C(=O)-[C:1]1-C-C-[C:2]-1"
product_smls = plot_rxn(pyrrolidine_to_cyclobutane, pyrrolidine_mol)
Reaction:
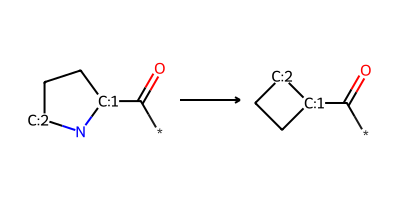
Reactant:
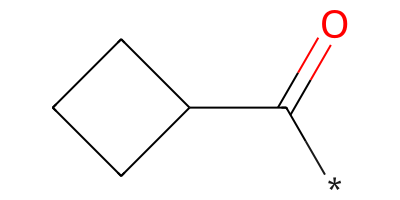
Distinct products:
*C(=O)C1CCC1
pyrrolidine_to_cyclobutane_modified = (
"[C:5](=O)-[C:1]1-[C:3]-[C:4]-[C:2]-N-1>>[C:5](=O)-[C:1]1-[C:3]-[C:4]-[C:2]-1"
)
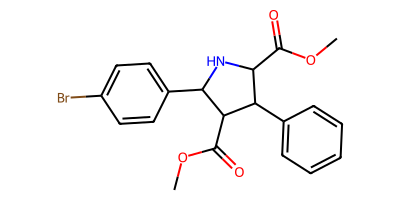
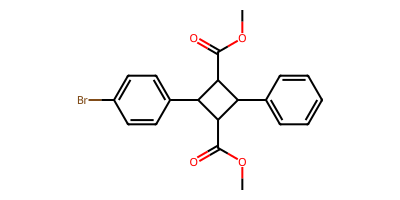
starting_smi = "c1cc(Br)ccc1C2NC(C(=O)OC)C(c3ccccc3)C(C(=O)OC)2"
starting_mol = Chem.MolFromSmiles(starting_smi)
product_smls = plot_rxn(pyrrolidine_to_cyclobutane_modified, starting_mol)
Reaction:
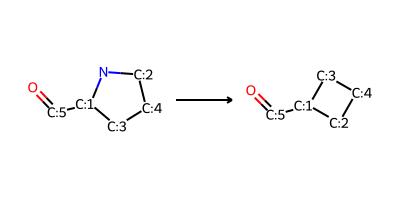
Reactant:
Distinct products:
COC(=O)C1C(c2ccccc2)C(C(=O)OC)C1c1ccc(Br)cc1
Converting nitroarenes into azepanes
This reaction is from Mykura et al., 2024.
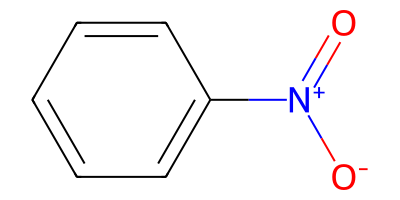
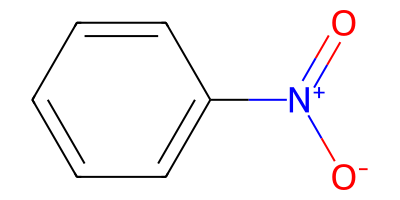
nitroarene = "O=[N+]([O-])c1ccccc1"
nitroarene_mol = Chem.MolFromSmiles(nitroarene)
nitroarene_mol
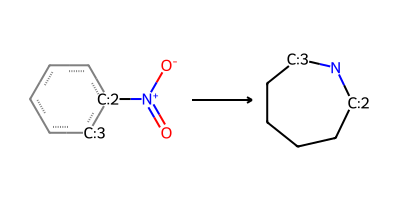
nitroarene_to_azepane = "O=[N+](-[O-])-[c:2]1[c:3]cccc1>>[C:2]-1-[N]-[C:3]-C-C-C-C1"
product_smls = plot_rxn(nitroarene_to_azepane, nitroarene_mol)
Reaction:
Reactant:
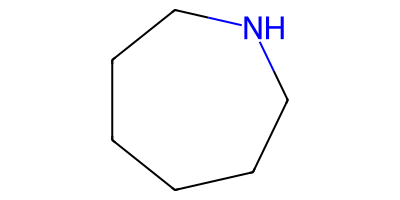
Distinct products:
C1CCCNCC1
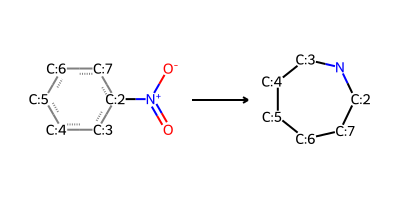
nitroarene_to_azepane_modified = "O=[N+](-[O-])-[c:2]1[c:3][c:4][c:5][c:6][c:7]1>>[C:2]-1-[N]-[C:3]-[C:4]-[C:5]-[C:6]-[C:7]1"
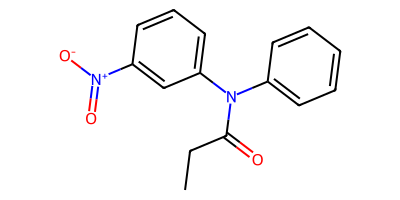
starting_smi = "CCC(=O)N(c1ccccc1)c3cccc([N+](=O)[O-])c3"
starting_mol = Chem.MolFromSmiles(starting_smi)
product_smls = plot_rxn(nitroarene_to_azepane_modified, starting_mol)
Reaction:
Reactant:
Distinct products:
CCC(=O)N(c1ccccc1)C1CCCNCC1
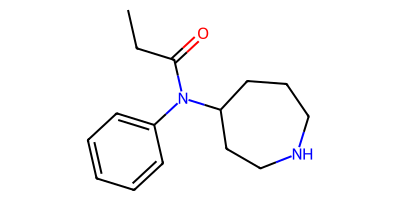
CCC(=O)N(c1ccccc1)C1CCCCNC1
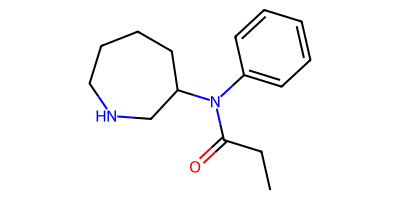
Phyo Phyo discusses the distinction between these two products and the factors favoring one over the other in the blog post How to Use Skeletal Editing in Drug Discovery: Key Benefits & Applications.
Update
Here’s a follow-up tutorial: Skeletal Editing with Reaction SMARTS and RDKit Atom Substitution